The unmapped chemical complexity of our diet
Abstract
Our understanding of how diet affects health is limited to 150 key nutritional components that are tracked and catalogued by the United States Department of Agriculture and other national databases. Although this knowledge has been transformative for health sciences, helping unveil the role of calories, sugar, fat, vitamins and other nutritional factors in the emergence of common diseases, these nutritional components represent only a small fraction of the more than 26,000 distinct, definable biochemicals present in our food—many of which have documented effects on health but remain unquantified in any systematic fashion across different individual foods. Using new advances such as machine learning, a high-resolution library of these biochemicals could enable the systematic study of the full biochemical spectrum of our diets, opening new avenues for understanding the composition of what we eat, and how it affects health and disease.
Main
The maxim of Jean Anthelme Brillat-Savarin, “Dites-moi ce que vous mangez et je vais vous dire ce que vous êtes”—‘you are what you eat’—remains as pertinent today, in the era of modern medicine, as it did in 1826. Indeed, the exceptional role of diet in health is well documented by decades of research in nutritional epidemiology, unveiling the role of nutrients and other dietary factors in cardiovascular disease, obesity, type 2 diabetes mellitus (T2DM) and other common diseases1. Yet, the bulk of our current understanding of the way food affects health is anchored in the 150 nutritional components that the United States Department of Agriculture (USDA) and other national databases track2,3, and these nutritional components represent only a subset of the total pool of definable biochemicals in the food supply (see Supplementary Discussion 1).
The dark matter of nutrition
Consider garlic, a key ingredient of the Mediterranean diet: the USDA quantifies 67 nutritional components in raw garlic, indicating that this bulbous plant is particularly rich in manganese, vitamin B6 and selenium4. However, a clove of garlic contains more than 2,306 distinct chemical components5,6—from allicin, an organosulfur compound responsible for the distinct aroma of the freshly crushed herb, to luteolin, a flavone with reported protective effects in cardiovascular disease7—which are listed in FooDB, a database representing the most comprehensive effort to integrate food composition data from specialized databases and experimental data. As of August 2019, FooDB records the presence of 26,625 distinct biochemicals in food8,9, a number that is expected to increase in the near future (see Supplementary Discussion 2). This exceptional chemical diversity could be viewed as the ‘dark matter’ of nutrition, as most of these chemicals remain largely invisible to both epidemiological studies, as well as to the public at large.
Where does this remarkable chemical diversity come from? Living organisms require a large number of biochemicals to grow and survive in their limited environments, well beyond the nutritional components that we humans need in our diet. From an evolutionary perspective, plants are characterized by a particularly rich chemical composition, mainly because they are unable to outrun their predators; their defence is occasionally mechanical (for example, through the development of spikes) but is predominantly chemical, exercised through smell, taste and appearance. These chemical defences require an extensive secondary metabolism that produces a wide range of flavonoids, terpenoids and alkaloids. Polyphenols—a highly studied group of chemicals believed to be responsible for the health effects of tea and other plants—are the product of that secondary metabolism. The number of secondary metabolites is estimated to exceed 49,000 compounds, indicating that the 26,000 chemicals currently assigned to food represent an incomplete assessment of the true complexity of the ingredients we consume10. Multiple environmental factors, from light to soil moisture, fertility and salinity, can influence the biosynthesis and accumulation of such secondary metabolites11. Humans and other animals who can hunt for the necessary food sources do not have the ability to synthesize many molecules our metabolism requires, like ascorbic acid or alpha-linolenic acid, necessitating a source for these essential nutrients.
Overall, an analysis of USDA and FooDB data confirms that plants as a group have the highest chemical diversity, with approximately 2,000 chemicals detected in most examples. Yet, 85% of these chemicals remain unquantified, meaning that while their presence has been detected or inferred, their concentration in specific food ingredients remains unknown (see Supplementary Discussion 2). With garlic, for example, FooDB reports the chemical concentration for just 146 chemical components; the remaining 2,160 chemicals listed in FooDB are not quantified5,6. We, therefore, raised the question as to whether the scientific literature contains valuable information on food composition beyond that currently compiled by food databases. Indeed, experimental and analytical projects focused on specific foods and foodborne chemicals are published on a daily basis, and only a small fraction of them inform databases. To unveil this potentially hidden knowledge, we developed a pilot project, FoodMine, that uses natural language processing to mine the full scientific literature for the purpose of comprehensively expanding all available scientific data on the biochemical composition of foods12.
FoodMine identified 5,676 papers from PubMed that potentially report on chemicals pertaining to the detailed chemical composition of garlic. After filtering this list using machine learning, we manually evaluated 299 papers, of which 77 reported 1,426 individual chemical measurements pertaining to garlic’s chemical composition. Our pilot project recovered more unique quantified compounds than are catalogued by the USDA and FooDB together (see Supplementary Discussion 3 and Supplementary Table 1). For example, diallyl disulfide is known to contribute to garlic’s smell and taste, and is implicated in the reported health benefits of garlic, as well as in garlic allergy13,14. Although FoodMine found multiple publications reporting on its concentration in garlic, the current databases do not offer quantified information for the compound. Furthermore, FoodMine identified information for 170 compounds that were not previously linked to garlic, either in the USDA or FooDB database (see Supplementary Discussion 3).
Taken together, we find that there is a wealth of exceptionally detailed information about food composition scattered across multiple literature sources. The current incompleteness in coverage within existing food composition databases is not due to a lack of interest in these chemicals or lack of efforts to map these chemical building blocks of food. Rather, it reflects the absence of systematic in-depth efforts to identify and catalogue the data scattered across multiple scientific communities and literature sources. As we discuss below, high-throughput tools required to scan the scientific literature and to overcome these limitations have emerged in the past several years. Mobilizing them could set the stage for an in-depth and systematic understanding of the ways by which our food affects health.
Health implications
The focus on a relatively limited group of nutritional components, including salt, sugar, protein and fat has been justified, given the important role each of them plays in health and disease. Yet, many documented health effects may be linked to untracked chemicals. Consider, for example, trimethylamine N-oxide (TMAO)15. Recent studies have found that patients with stable coronary heart disease had a fourfold greater risk of dying from any cause over the subsequent five years if they had high blood levels of TMAO16.
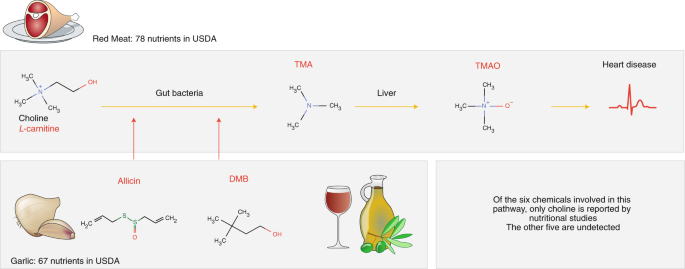
While TMAO and its precursor trimethylamine (TMA) naturally occur in fish and milk, important sources of TMAO in the Western diet are L-carnitine and choline, both of which are found in red meat17,18. These molecules are metabolized by gut bacteria into TMA, which is then converted in the liver to TMAO18 (Fig. 1). The Mediterranean diet19, which regularly pairs red meat with fresh garlic, derives some of its known health benefits from allicin20, which blocks TMA production in the gut, ultimately lowering the TMAO concentration in plasma. Taken together, there are at least six distinct biochemicals in our diet involved in the TMAO pathway: L-carnitine, choline, TMA, TMAO, allicin and 3,3-dimethylbutan-1-ol (DMB). Yet, only one of them, choline, is tracked and quantified in nutritional databases21. The remaining five, despite the key roles they play in health, are effectively nutritional dark matter (Fig. 1).
Animal products contain L-carnitine, choline and choline-contributing compounds21. These molecules are metabolized by gut bacteria into trimethylamine (TMA), which is converted in the liver to trimethylamine-N-oxide17 (TMAO), a compound linked to coronary events16. Garlic, extra-virgin olive oil and red wine, staple ingredients of the Mediterranean diet, reduce the production of TMAO through allicin and 3,3-dimethylbutan-1-ol (DMB), compounds that block TMA production by gut bacteria. Of the six biochemical compounds involved in this pathway, only one, choline, is tracked in food by the USDA. The other compounds are part of the nutritional ‘dark matter’ (in red).
Overall, 37 nutritional components of garlic can be linked to diseases according to the Comparative Toxicogenomics Database (CTD)22. Indeed, garlic carries vitamins B1, B6 and C, and the minerals manganese, copper, selenium and calcium—nutrients whose deficiency or excess have been linked to disease such as T2DM, Parkinson’s disease and cardiomyopathies. These links confirm the important role the currently tracked nutrients play in health23 (see Supplementary Discussion 4). At the same time, the CTD reveals that 485 of the currently unquantified chemicals in garlic can also be linked to multiple therapeutic effects, like the protective action of allicin in cardiovascular disease24,25 discussed above.
There is a remarkable parallel between pre-genome biology and our current understanding of the health implications of diet. Indeed, in the 1980s, detractors of the Human Genome Project insisted that only the coding regions, representing 1.4% of all base pairs in our DNA, are worth the cost of decoding, labelling the remaining 98.6% ‘junk DNA’. Yet, today it is estimated that 66% of disease-carrying variants are, in fact, in these non-coding regions. Similarly, today the 150 nutritional components tracked in food composition tables represent about 0.5% of the 26,625 chemical compounds documented in food. The health implications of these nutritional components are well studied. Yet, more than 99% of the biochemicals present in food, many of which play a role in health and disease, are untracked by national databases, with the health implications of this largely unexplored nutritional dark matter remaining largely unknown. The absence of information on these untracked biochemicals could be responsible for inconsistencies in, and the irreproducibility of, published results as well as for missing health effects, and can also create spurious associations that are not replicable by meta-analysis26.
Advances in network medicine27,28,29,30,31—a post-genome discipline that emphasizes the role of comprehensive molecular interactions (comprising a molecular interaction network, or interactome) in the prevention and treatment of disease—could help us to systematically unveil the mechanistic role of the wide array of molecules found in our diet. Consider, for example, the polyphenol (–)-epigallocatechin 3-O-gallate (EGCG), an abundant biochemical compound in green tea, with potential therapeutic effects in T2DM. Network-based metrics reveal a proximity between 52 human proteins targets of EGCG32 and 83 proteins associated with T2DM30,33. This offers multiple mechanistic pathways by which to account for the relationship between green tea consumption and its many reported effects on health and disease risk34,35,36, and its glucose-lowering effects observed using in vitro and in vivo models37,38. Unveiling the nutritional dark matter could open up new strategies for discovering the wide array of molecular mechanisms through which food affects health, helping us understand how to use food as therapy, and to aid the identification of food biochemicals with direct therapeutic impact.
Food affects our health through multiple molecular mechanisms: some chemicals serve as a direct source of intermediates for human metabolism, while others, such as polyphenols, play a regulatory role. Yet many food molecules also feed the microbiome in our gut, which metabolizes these compounds into other species that can be further transformed by mammalian metabolism (such as TMA and TMAO)39,40. Tracking the full chemical composition of the specific ingredients is also unavoidable if we wish to gain a better understanding of the many ways by which the microbiome responds to the vast diversity of our diet, and how best to alter the microbiome for therapeutic purposes.
Mapping out the foodome
The current incomplete chemical profiling of food poses a number of fundamental scientific and methodological challenges, limiting our ability to systematically explore the health implications of our diet. Yet, the multiple ‘known unknowns’ of nutrition offer a potential roadmap by which to address them. Indeed, a systematic mapping of the complete chemical composition of the food we consume, although costly, is feasible, and could be greatly accelerated by recent advances in the use of big data and artificial intelligence.
For example, the remarkable governmental and community efforts behind databases like the USDA3,41,42, FooDB43, Frida2, PhenolExplorer44 and eBasis45 have already resulted in a wealth of information on food composition. Rapid advances in metabolic reconstructions and biochemical modelling enable us to infer specific pathways from the genome, and through machine learning we can combine metabolic pathway information with the existing food composition databases in a systematic fashion, potentially elucidating the missing chemicals. Indeed, the closer two ingredients are on the phylogenetic tree46, the more similar is their expected metabolic pathway structure and biochemical composition. Machine learning is ideally suited to combine the known chemical composition of chosen food ingredients over different taxonomical branches with the list of orthologous enzymes in sequenced organisms; the missing chemical information can then be elucidated by learning the appropriate distance metric47,48 between organisms and clustering correlated groups of pathways and biochemicals49. Such efforts, taking full advantage of existing knowledge, could offer experimentally verifiable predictions about the missing chemicals and their concentration.
Yet, part of the challenge is experimental: the time-consuming, low-throughput, structural chemical tools (spectroscopic methods, nuclear magnetic resonance, mass spectrometry and so on) may need to be fundamentally reengineered into high-throughput methods that can scan food with sufficient chemical resolution and sensitivity, helping to catalogue the presence and the concentration of the vast array of currently unquantified chemical compounds in the food supply. Such efforts are complemented by ‘foodomics’, a movement aiming to bring omics-technology to the systematic exploration of food50,51.
Cooking and food processing alter the chemical composition of food, adding chemicals that are absent in raw ingredients and transforming others, from emulsifiers to new lipids. Some of these changes have well documented health implications, like the presence of acrylamide (a carcinogenic compound) in fried and baked goods and in coffee. While the impact of food processing on basic nutritional components is well studied, little is known about the impact of processing on the thousands of chemicals found in the nutritional dark matter. Equally important, we must account for the numerous toxins added to food during cooking, preservation and packaging, or accumulated in food according to the environmental production conditions, and their effect on health, such as the well documented toxicity of highly reactive aldehydes or of persistent organic pollutants52.
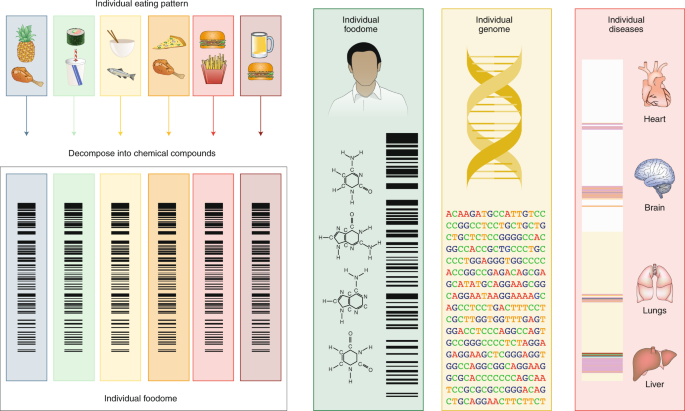
For nutrition to compete with genetics in accuracy, reach and impact, we must organize the information on eating patterns to fit the big-data platform that fuels advances in this digital age of biomedicine. Indeed, our eating patterns are digital—each of us consumes a weighted subset of chemicals found in the food supply. The precise subset of chemicals to which each individual is exposed defines that person’s individual nutritional-chemical ‘barcode’, or his or her ‘foodome’ (Fig. 2). The determinants of this personal foodome are complex, from food supply to personal choices, and are modulated by geography, culture and socio-economic status. Efforts to ensure the traceability53 of food, allowing us to track the source and production of the raw material introduced into the food chain, together with the environmental and processing conditions modulating the individual foodome, will also greatly enhance future research in this arena. Our ability to track the nutritional–chemical barcode of each individual, and correlate it with individual genetic variations and health history, could help merge nutrition with a precise digital and statistical platform similar to that which fuelled the spectacular advances in genomics54,55. Such a platform could help us scan systematically for novel causal mutation–chemical–health associations that are largely invisible to current hypothesis-driven research in nutrition.
Our daily eating patterns define a unique biochemical barcode, representing a high-resolution description of each person’s individual biochemical exposure through his or her diet, or individual foodome. To assess the individual foodome in a reliable fashion, we can take advantage of the smartphone revolution and collect daily food diaries59 via image capture. Combined with genomics and disease histories, access to this full biochemical palette could help us expand the widely used genome-wide-association-study-based tools to account for the biochemical composition of our eating patterns, and systematically unveil the linkages between specific food biochemicals, genome variations and health.
To appreciate the transformative potential of a deeper quantitative understanding of the nutritional dark matter, we must realize that our genetic predispositions to specific phenotypes and pathophenotypes can conceivably be modified by these food-based molecules. Indeed, while we cannot currently change the genetic basis for disease, we regularly modulate the activity of our subcellular networks through the food we eat, diminishing the impact of some mutations and enhancing the role of others. This differential modulation of subcellular networks explains why individuals with strong genetic predispositions to heart disease can lower the chance of developing the disease by up to 70% with proper lifestyle choices56, within which dietary changes play a dominant role56,57. This finding implies that an accurate mapping of our full chemical exposure through our diet could lead to actionable information to improve health. Recent trends in nutrition research, aiming to explore the synergies, competitions and interactions among the entire matrix of what constitutes a food product, increasingly acknowledge the complexity of the problem, and the need for new tools to address it58. We must embrace this irreducible complexity to be able to integrate changes in the food supply, the role of the microbiome and personalized dietary patterns, so that we can eventually offer individually tailored food-based therapies and appropriate lifestyle choices for disease prevention and lifespan optimization.
References
- 1.Willett, W. Nutritional Epidemiology (Oxford Univ. Press, 1990).
No comments:
Post a Comment